What to know
- Below are confirmed norovirus outbreaks submitted to CaliciNet and updated monthly.
- Participating public health laboratories electronically submit genotype information for norovirus and basic epidemiology data from norovirus outbreaks to the CaliciNet database.
- The data on norovirus strains are compared with existing norovirus sequences in the CaliciNet database.
Genotype distribution of norovirus outbreaks
September 1, 2024 – December 31, 2024 (n=56)
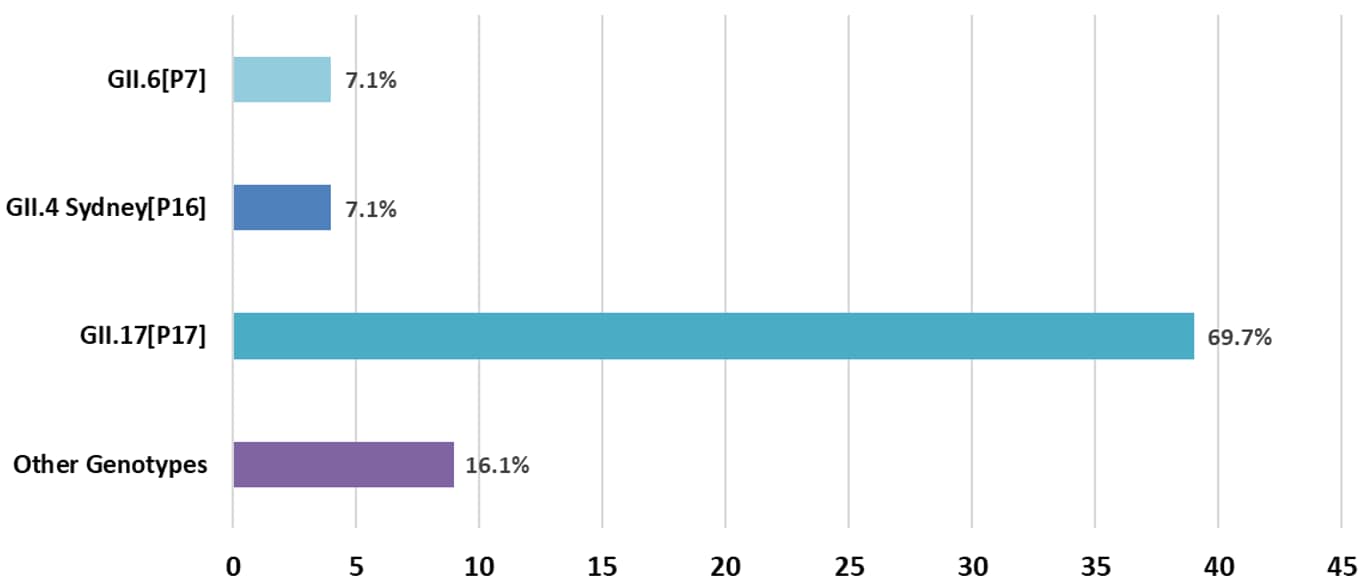
The Other Genotypes category includes genotypes from outbreaks that each make up less than 5% of the total number of outbreaks. These genotypes are: GI.2[P2], GI.3[P3], GI.3[P13], GI.5[P5], GI.7[P7], GII.3[P12], GII.7[P7].
Data Table: September 1, 2024 – December 31, 2024 (n=56)
| Genotype | Number of Outbreaks |
|---|---|
| GII.17[P17] | 39 |
| GII.4 Sydney[P16] | 4 |
| GII.6[P7] | 4 |
| Other Genotypes | 9 |
Data Table: September 1, 2023 – August 31, 2023 (n=309)
| Genotype | Number of Outbreaks |
|---|---|
| GII.17[P17] | 109 |
| GII.4 Sydney[P16] | 84 |
| GII.6[P7] | 29 |
| GI.5[P5] | 26 |
| Other Genotype | 61 |
Dual typing for norovirus
Dual typing for norovirus is being implemented by laboratories participating in CaliciNet.
Dual typing is RT-PCR amplification of a partial region of both the polymerase gene and the capsid gene (region B-C) in a single reaction for genogroup I or genogroup II noroviruses.
